Research on possible CoVID-19 drug compounds using docking studies
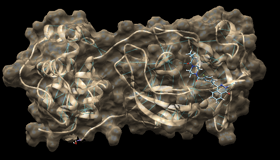
During the tough pandemic times, I had a bit more time at hand and decided to provide a little bit of help to the CoVID research community while also learning more about the packages and methodologies involved in the process of docking compounds and repurposing drugs. Repurposing has been viewed as a part of the primary analysis in any drug discovery pipeline since if the currently apporved drugs work better than a new molecule in in-silico studies, it also saves a lot of time and money for the process of approving a new drug while making the prescription more readily avaiable to the medical facilities. I did a simple python docking procedure for a select case of drugs and visualized their docking scores while on the other project, I prepared a CoVID 19 Drug Repurposing Analysis tool using R to use meta analyze the drug datatbase to find best repurpopsing compound. Basic docking analysis was performed using the method listed in https://github.com/omarwagih/covid19-docking with the protein list obtained after meta analysis of various molecules. This project’s paper is under review currently.